Support Vector Machines (SVM)
Introduction
Support Vector Machines (SVM) are powerful supervised learning models used for classification and regression tasks. SVMs work by finding the hyperplane that best separates the classes in the feature space. R provides excellent packages like e1071 and caret to build and evaluate SVM models.
Key Steps in Building an SVM in R
Data Preparation: Cleaning and preparing data for analysis.
Model Training: Building the SVM model.
Model Evaluation: Assessing the model’s performance.
Prediction: Using the trained model to make predictions on new data.
Example: Predicting Species with SVM
Load and Prepare the Data
Code
# Load the data
data <- iris
# Split the data into training and testing sets
set.seed(123)
train_index <- createDataPartition(data$Species, p = 0.8, list = FALSE)
train_data <- data[train_index, ]
test_data <- data[-train_index, ]Train the SVM Model
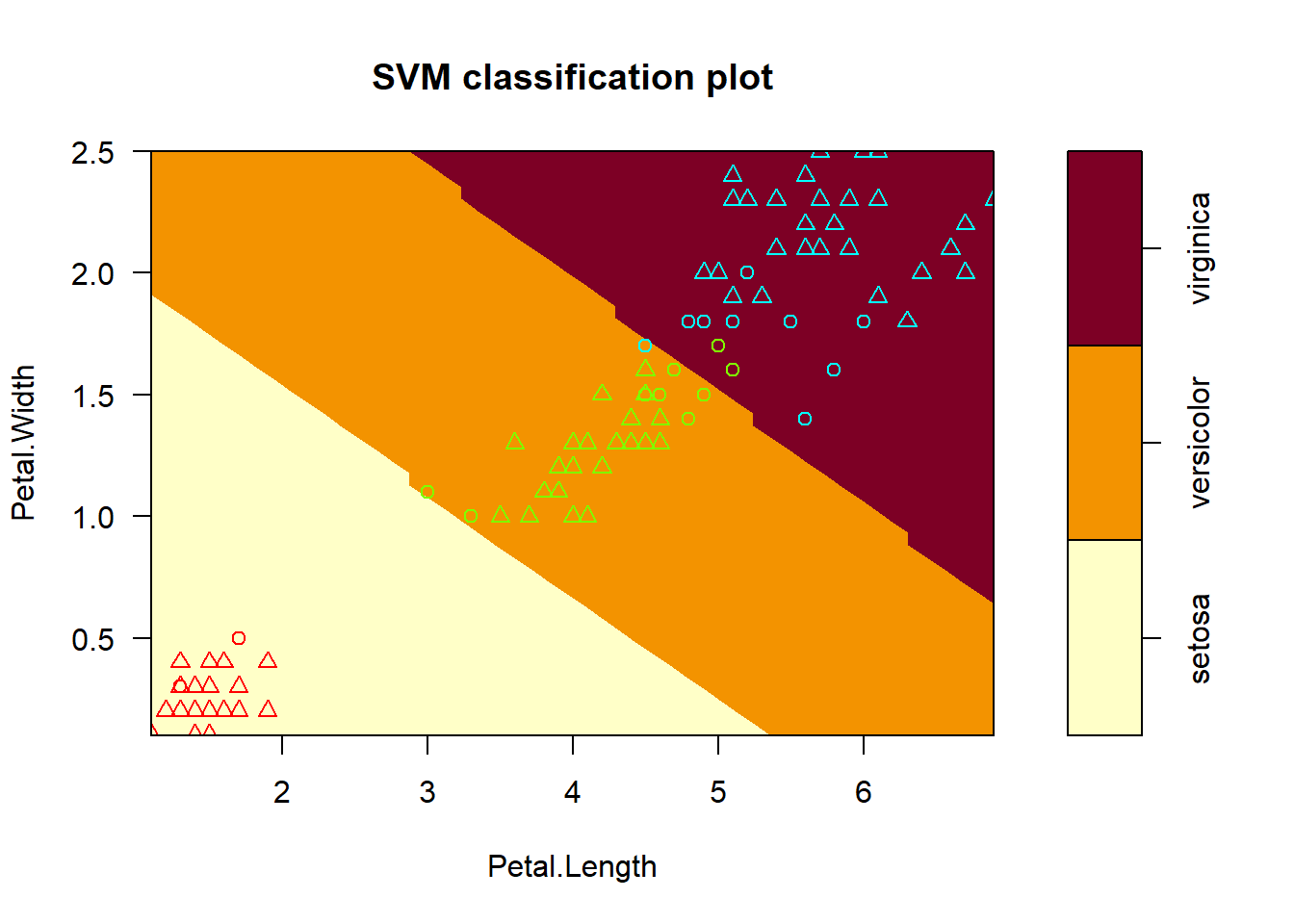
Ploting the Model
Evaluate the Model
Assess the model’s performance on the testing set.
Code
# Make predictions on the testing set
predictions <- predict(model, newdata = test_data)
# Create a confusion matrix
conf_matrix <- confusionMatrix(predictions, test_data$Species)
print(conf_matrix)Confusion Matrix and Statistics
Reference
Prediction setosa versicolor virginica
setosa 10 0 0
versicolor 0 10 1
virginica 0 0 9
Overall Statistics
Accuracy : 0.9667
95% CI : (0.8278, 0.9992)
No Information Rate : 0.3333
P-Value [Acc > NIR] : 2.963e-13
Kappa : 0.95
Mcnemar's Test P-Value : NA
Statistics by Class:
Class: setosa Class: versicolor Class: virginica
Sensitivity 1.0000 1.0000 0.9000
Specificity 1.0000 0.9500 1.0000
Pos Pred Value 1.0000 0.9091 1.0000
Neg Pred Value 1.0000 1.0000 0.9524
Prevalence 0.3333 0.3333 0.3333
Detection Rate 0.3333 0.3333 0.3000
Detection Prevalence 0.3333 0.3667 0.3000
Balanced Accuracy 1.0000 0.9750 0.9500Prediction on New Data
Use the tuned model to make predictions on new data.
Code
1 2
setosa virginica
Levels: setosa versicolor virginicaSVM Regression
Code
crim zn indus chas nox rm age dis rad tax ptratio b lstat
1 0.00632 18 2.31 0 0.538 6.575 65.2 4.0900 1 296 15.3 396.90 4.98
2 0.02731 0 7.07 0 0.469 6.421 78.9 4.9671 2 242 17.8 396.90 9.14
3 0.02729 0 7.07 0 0.469 7.185 61.1 4.9671 2 242 17.8 392.83 4.03
4 0.03237 0 2.18 0 0.458 6.998 45.8 6.0622 3 222 18.7 394.63 2.94
5 0.06905 0 2.18 0 0.458 7.147 54.2 6.0622 3 222 18.7 396.90 5.33
6 0.02985 0 2.18 0 0.458 6.430 58.7 6.0622 3 222 18.7 394.12 5.21
medv
1 24.0
2 21.6
3 34.7
4 33.4
5 36.2
6 28.7Splitting the data
Fitting the model
Code
s <- svm(medv ~ ., data=train)
s
Call:
svm(formula = medv ~ ., data = train)
Parameters:
SVM-Type: eps-regression
SVM-Kernel: radial
cost: 1
gamma: 0.07142857
epsilon: 0.1
Number of Support Vectors: 274Predictions
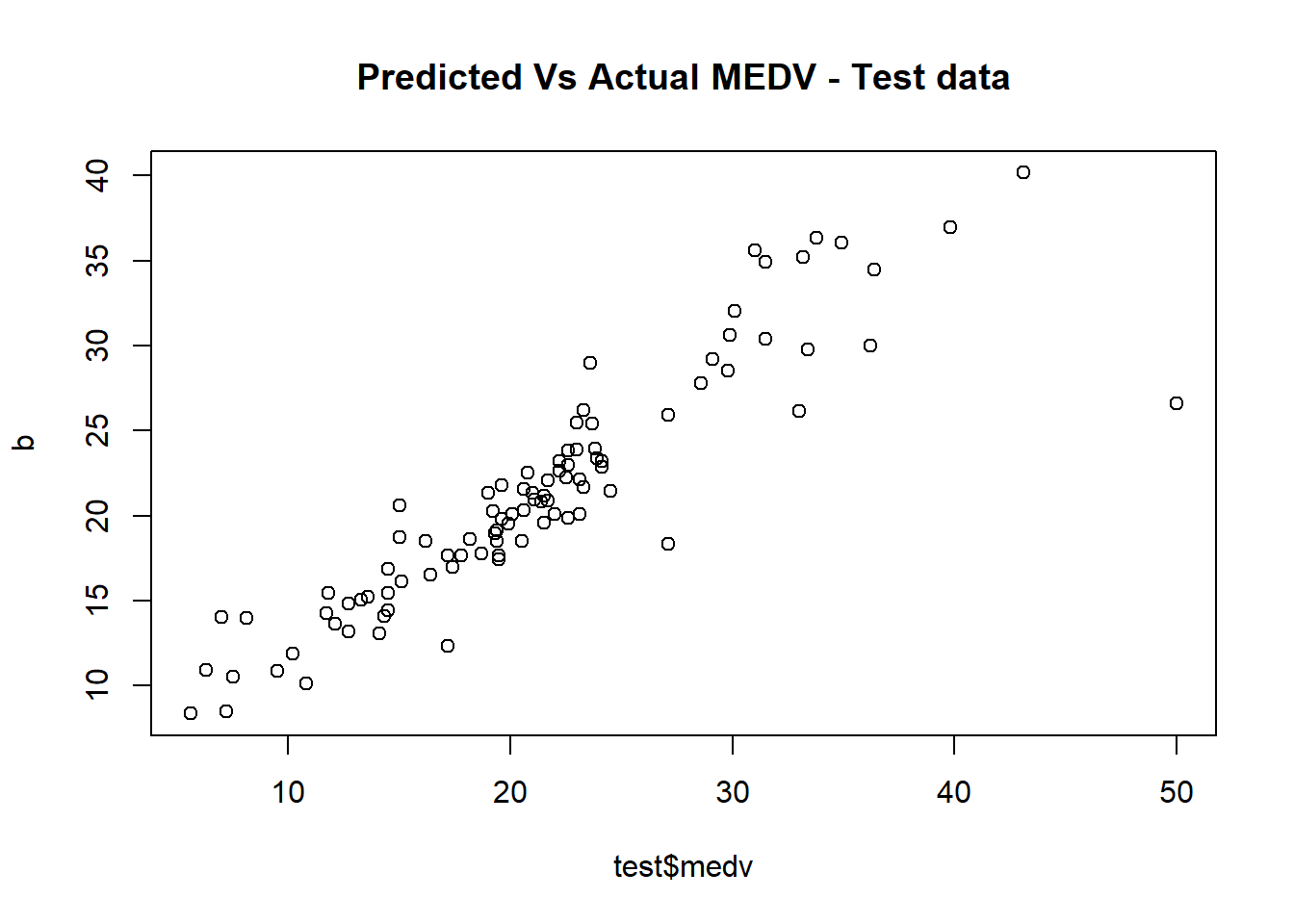
Scatter plot of Actual vs Predicted
Code
plot(b~ test$medv, main = 'Predicted Vs Actual MEDV - Test data')