Introduction
Breast cancer remains a significant global health concern, affecting millions of individuals worldwide and posing formidable challenges to healthcare systems. Timely and accurate diagnosis is paramount in guiding treatment decisions and improving patient outcomes. This project delves into the realm of predictive modeling specifically focused on breast cancer diagnosis. By harnessing the power of machine learning and leveraging a rich dataset comprising clinical and diagnostic features, this endeavor aims to develop a robust predictive model capable of discerning between malignant and benign tumors with high accuracy.
Variables
Diagnosis: This categorical variable indicates the diagnosis outcome, with “M” representing malignant tumors and “B” representing benign tumors. It serves as the target variable for prediction.
Radius_mean, Texture_mean, Perimeter_mean: These variables represent the mean values of the radius, texture, and perimeter of the tumor cells, respectively, measured from diagnostic images.
Area_mean, Smoothness_mean, Compactness_mean, Concavity_mean, Concave_points_mean: These variables capture additional morphological characteristics of tumor cells, such as the area, smoothness, compactness, concavity, and number of concave points in the cell nuclei, averaged across the diagnostic images.
Symmetry_mean, Fractal_dimension_mean: These variables describe symmetry and fractal dimension features of tumor cells, providing insights into their structural complexity and irregularity.
Radius_se, Texture_se, Perimeter_se, Area_se, Smoothness_se, Compactness_se, Concavity_se, Concave_points_se, Symmetry_se, Fractal_dimension_se: These variables represent the standard error (SE) of the corresponding morphological features, providing information about the variability or uncertainty in their measurements.
Radius_worst, Texture_worst, Perimeter_worst, Area_worst, Smoothness_worst, Compactness_worst, Concavity_worst, Concave_points_worst, Symmetry_worst, Fractal_dimension_worst: These variables denote the “worst” or largest values of the respective morphological features observed in the tumor cells, providing insights into the most severe manifestations of tumor characteristics.
Required packages
Data Importation and Preprocessing
Exploratory Data Analysis
Code
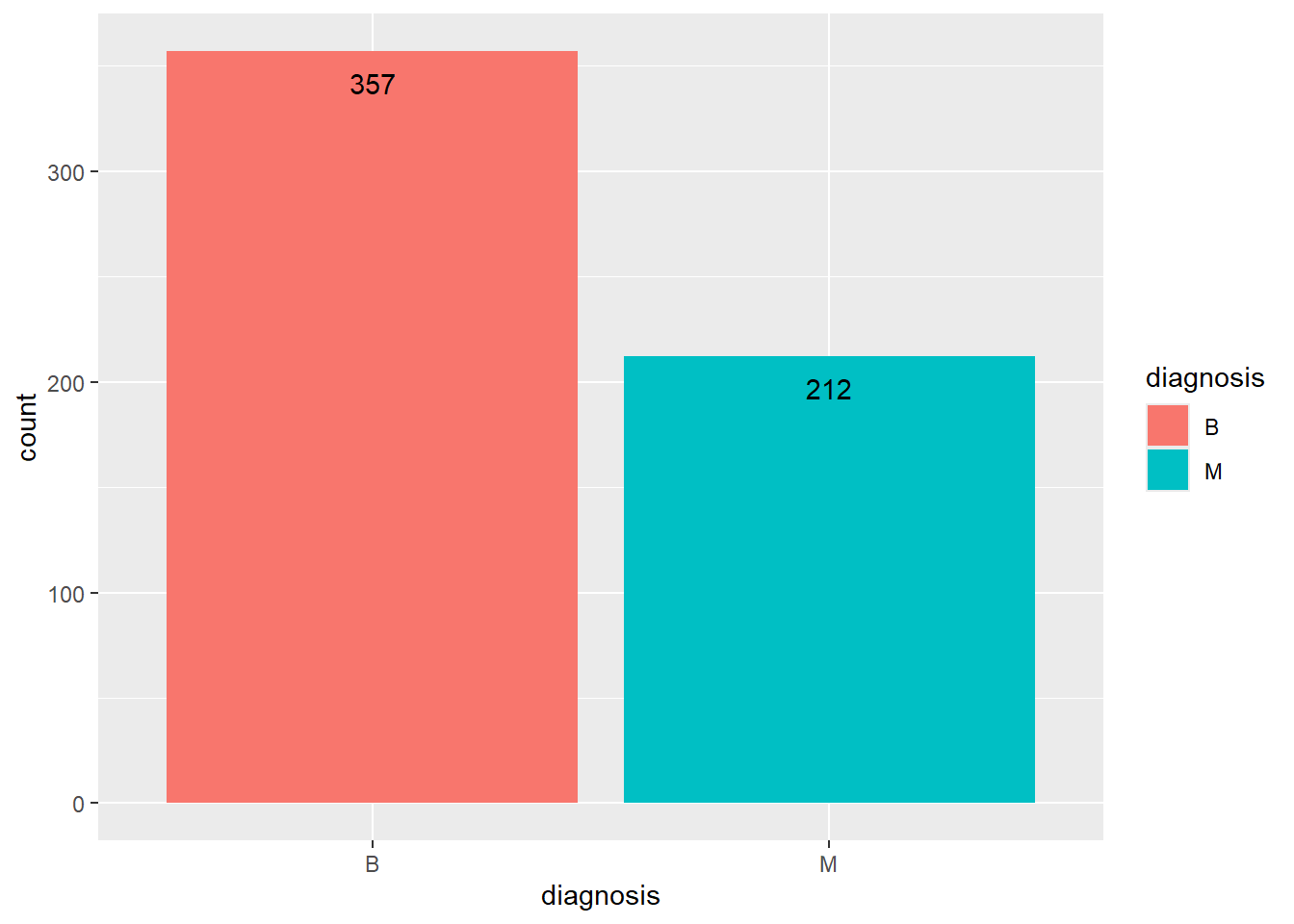
“B” indicates benign tumors. “M” indicates malignant tumors.
There are 357 instances of benign tumors (labelled as “B”). There are 212 instances of malignant tumors (labelled as “M”).
Checking for Dublicates
Code
cancer %>% duplicated() %>% unique()[1] FALSEThere are no dublicates
Balancing the Data by Oversampling
Code
B M
357 357 The data is now balanced
Spliting the Data into Training and Testing
Code
set.seed(123)
split <- initial_split(cancer, prop = 0.8, strata = diagnosis)
train <- training(split)
test <- testing(split)
tabyl(cancer$diagnosis) cancer$diagnosis n percent
B 357 0.5
M 357 0.5Code
tabyl(train$diagnosis) train$diagnosis n percent
B 285 0.5
M 285 0.5Code
tabyl(test$diagnosis) test$diagnosis n percent
B 72 0.5
M 72 0.5Building a Randomforest Classification Model
Random Forest
570 samples
30 predictor
2 classes: 'B', 'M'
No pre-processing
Resampling: Bootstrapped (25 reps)
Summary of sample sizes: 570, 570, 570, 570, 570, 570, ...
Resampling results across tuning parameters:
mtry Accuracy Kappa
2 0.9654780 0.9308282
16 0.9582624 0.9163916
30 0.9527224 0.9052925
Accuracy was used to select the optimal model using the largest value.
The final value used for the model was mtry = 2.Code
plot(m1)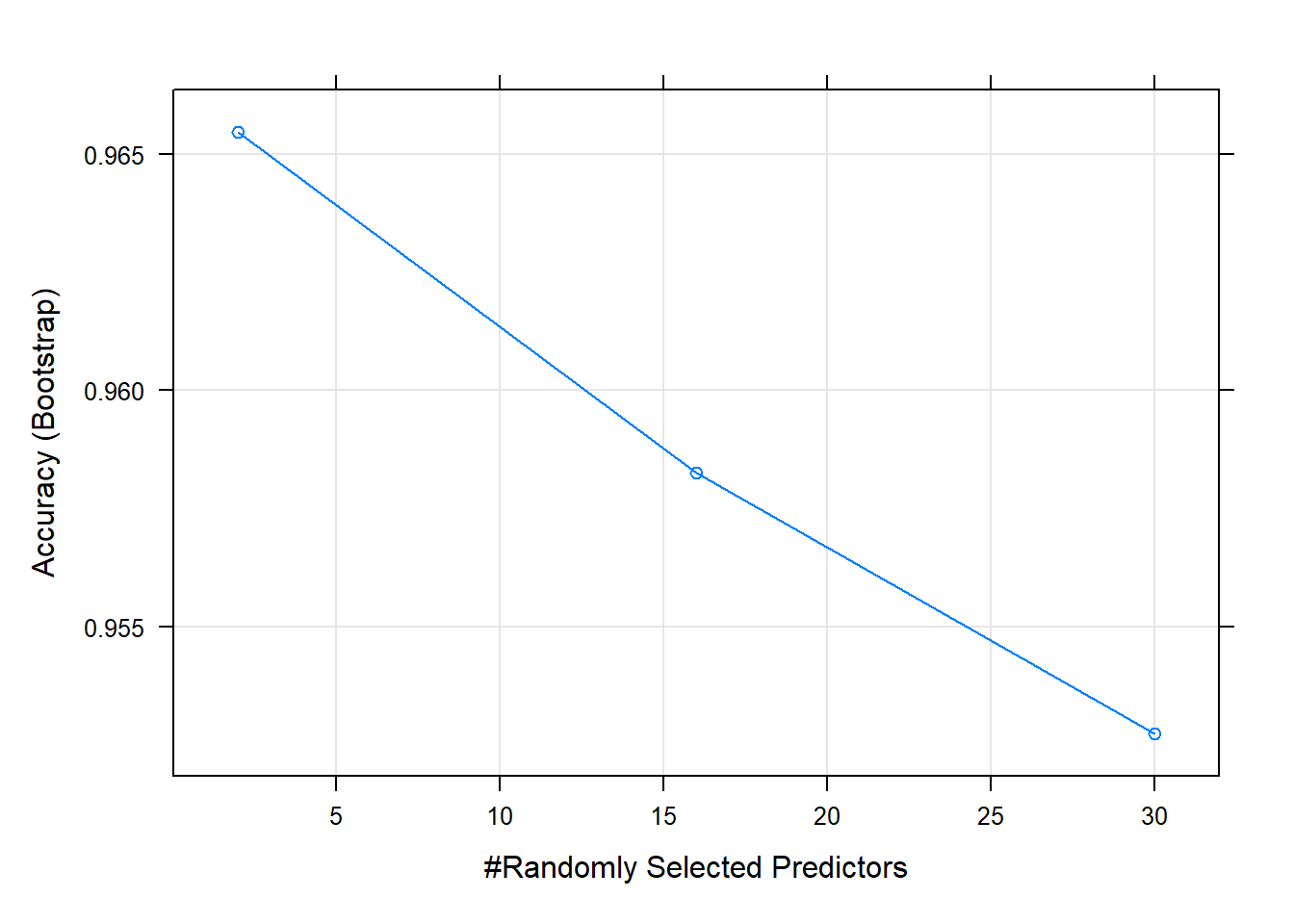
Variable Importance
Code
v<- varImp(m1)
vrf variable importance
only 20 most important variables shown (out of 30)
Importance
perimeter_worst 100.00
radius_worst 96.34
concave_points_worst 91.77
texture_worst 88.20
area_worst 85.11
concave_points_mean 80.29
concavity_worst 77.45
area_mean 75.47
perimeter_mean 72.75
radius_mean 69.84
area_se 69.81
texture_mean 68.16
concavity_mean 67.28
perimeter_se 61.78
smoothness_worst 59.77
radius_se 58.80
compactness_worst 51.80
symmetry_worst 50.13
fractal_dimension_worst 47.30
fractal_dimension_mean 43.47Code
plot(v)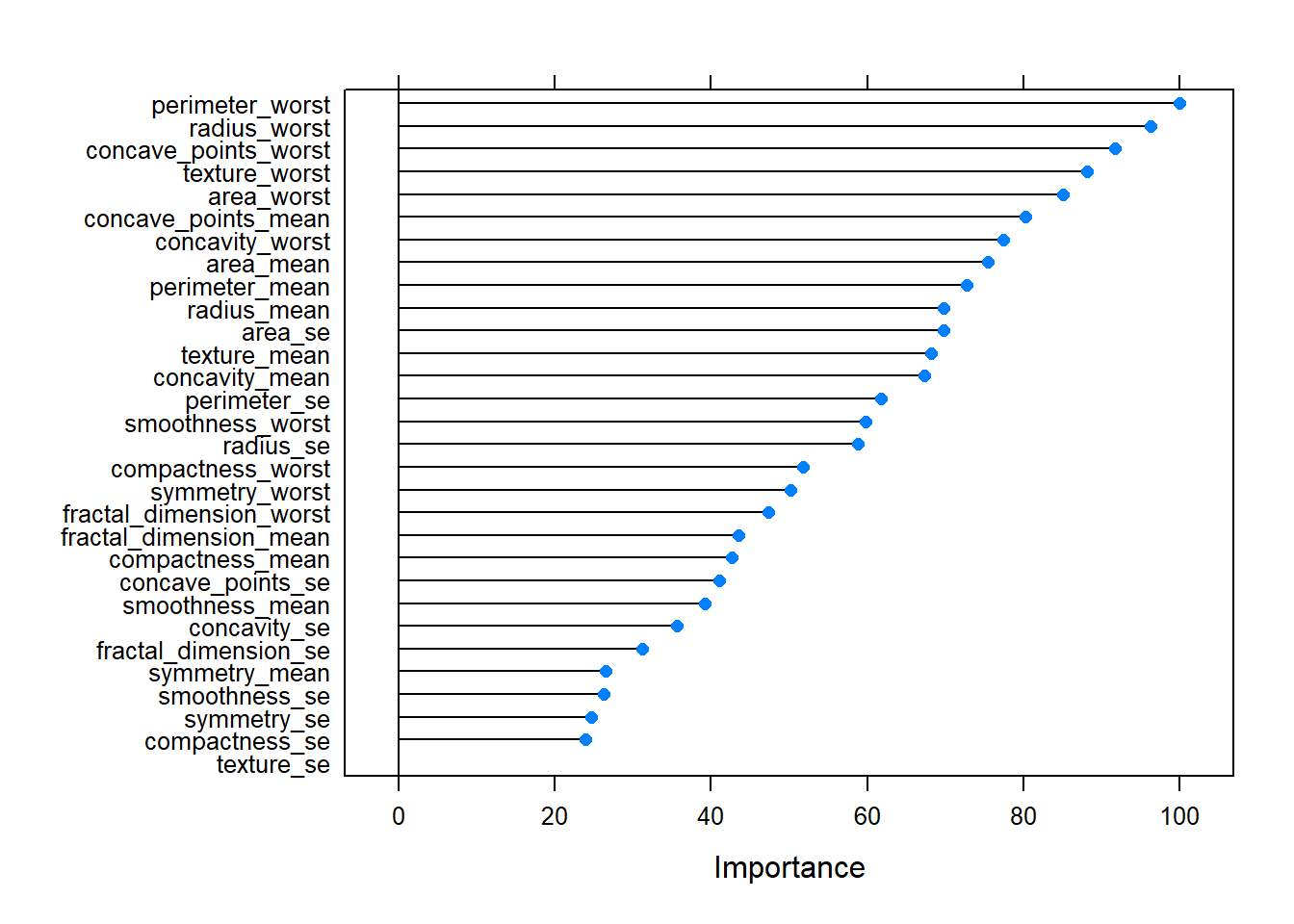
These values represent the importance of each predictor variable in contributing to the accuracy of the random forest model. Higher values suggest that the variable is more important in predicting the target variable (in this case, the diagnosis of breast cancer).
Predicting Using the Model
[1] B B B B B B
Levels: B MThe first five patients in the test data are predicted not to have cancer(Benign tumor)
Testing the Model Accuracy
Code
confusionMatrix(data = test$prediction, reference = test$diagnosis)Confusion Matrix and Statistics
Reference
Prediction B M
B 70 0
M 2 72
Accuracy : 0.9861
95% CI : (0.9507, 0.9983)
No Information Rate : 0.5
P-Value [Acc > NIR] : <2e-16
Kappa : 0.9722
Mcnemar's Test P-Value : 0.4795
Sensitivity : 0.9722
Specificity : 1.0000
Pos Pred Value : 1.0000
Neg Pred Value : 0.9730
Prevalence : 0.5000
Detection Rate : 0.4861
Detection Prevalence : 0.4861
Balanced Accuracy : 0.9861
'Positive' Class : B
Table of Predicted Against the Actual values
Explaining the 1st 3 Prediction
Code
library(lime)
e <- lime(test[1:3,], m1, n_bins = 4)
explanation <- explain( x = test[1:3,],
explainer = e,n_labels = 1,
n_features = 4)
b <- plot_features(explanation)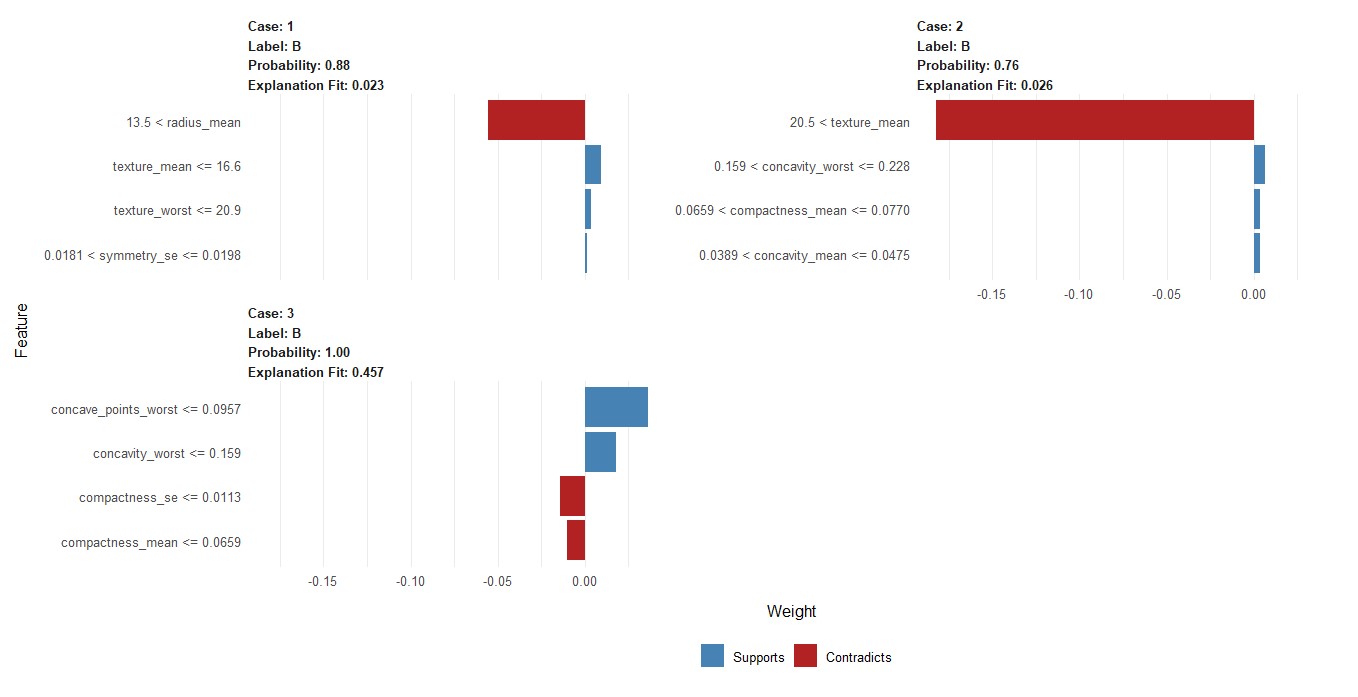
Limitation of the Model
Feature Selection: The predictive power of the model heavily relies on the choice of input features. While the current model considers various features such as radius, texture, perimeter, area, and others, there might be other relevant features not included in the analysis. Incorporating additional features or exploring feature engineering techniques could potentially enhance the model’s performance.
Model Interpretability: Random Forest models, like the one used here, are considered “black box” models, meaning they provide accurate predictions but offer limited interpretability. Understanding how individual features contribute to the model’s predictions can be challenging. Techniques such as feature importance analysis provide some insight but may not fully explain the underlying mechanisms driving the predictions.
External Factors: The model’s predictions may be influenced by factors not captured in the dataset, such as patient demographics, genetic factors, or environmental variables. Failing to account for these external factors could introduce bias or limit the model’s predictive accuracy in real-world applications.
Data source
Kaggle: link: https://www.kaggle.com/datasets/yasserh/breast-cancer-dataset